INTRODUCTION.
Colorectal cancer occupies the 4th place by the rate of lethality in patients. Annually, more than 1 million new cases and 715,000 lethal outcomes are registered [1, 2]. In Russia, during the past decade, the occurrence rate of colorectal cancer increased by 49.5%. The lethality rate reaches 40% within the first year because of the late diagnostics [3]. Some authors report that the rate of metastasis in patients with colorectal cancer exceeds 50%, and 5-year survival without specific treatment is not more than 2%. Such figures indicate an acute necessity in the implementation of new highly-specific diagnostic and prognostic molecular markers of colorectal cancer. Among the known tumor antigens, cancer/testis antigens (CTAs) that are expressed in tumors of different histologic origin and that are not expressed in normal tissues (except for testis, placenta, and brain) should be highlighted [4].
Presently, the discovered 276 C/T antigens are united in 138 families, 7 of which are well-described and studied (MAGE-A, -B, BAGE, GAGE, SSX, LAGE, MAGE-C) [5]. Polypeptides CTA in combination with HLA1 (Human Leukocyte Antigens 1, molecules of the major histocompatibility complex (MHC) of class II) or HLA2 (Human Leukocyte Antigens 2, molecules of the major histocompatibility complex (MHC) of class II) appear in the immune system, which causes cytotoxic and humoral responses [6]. Each type of tumor tissue has a highly-specific profile of C/T gene expression. The use of CTA as a molecular tumor marker or target for immune therapy requires the analysis of the respective databases or molecular-genetic studies for the identification of the most specific CTA [1, 7].
In patients with colorectal cancer, the characteristic of the expression of C/T genes is incomplete, especially, for the Russian population [8]. The first study on CTA in 54 patients with colorectal cancer performed in 1996 (expression of mRNA MAGEA-1; -2; -3) revealed the hyperexpression of MAGEA-1 in 30%, MAGEA-2 – in 28% and MAGEA-3 in 20% of cases. The expression of mRNA CTA was more often increased in patients with metastatic colorectal cancer [1]. Probably, aggressive forms of colon cancer primarily express MAGE family genes, which can contribute to the development of metastases. The degree of the expression of these genes or expression at least of one gene from this family in patients with the primary tumor of the colon is understudied in comparison with their expression in the metastases to the liver, which limits the conclusions that can be made based on these data [1, 9].
The published data on the expression of CTA in colon tumors show that certain representatives of 138 families of CTA have significant potential in the diagnostics and prognosis of metastases development in colon tumors [1].
The study was aimed to evaluate the transcription pr of C/T genes for the identification of CTA that are highly specific for colon tumors in patients with metastatic (T1-3N1-2M0) and non-metastatic (T1-3N0M0) cancer.
MATERIALS AND METHODS.
The study included operative biopsy samples of normal and tumor tissues of the colon obtained from 24 patients (48 samples) that underwent treatment at the Rostov-on-Don Scientific Research Institute of Oncology in 2016-2018. The study was approved by the local ethical committee. Each patient signed a form of informed consent for participation in the study. Tissue samples were instantly frozen in liquid nitrogen for the delivery to the lab [7].
Tissue fragments were homogenized in the lysing buffer that contained guanidinium thiocyanate, sodium citrate, sarcosil, and mercaptoethanol. The isolation of the total RNA from the tissue lysate was performed by the method of phenol-chloroform extraction [4, 10]. Genome DNA was isolated from the total RNA by deoxyribonuclease-1 drugs [4, 11]. For the evaluation of the quality of the obtained RNA specimen, the authors performed electrophoresis in a 2% agarose gel (the intensity of the stripes 18S and 28S in the ratio 1:1 indicated an acceptable quality of RNA for further study) [12]. The synthesis of cDNA was performed using commercial kits Reverta-L (Interlabservice, Russia) according to the manufacturer’s instructions [4, 7, 11]. The method of real-time quantitative PCR (RT- qPCR) was used for the evaluation of the relative expression of 16 genetic loci: MAGE-A1,-A2, -A3, -A4, -B1, -B2, GAGE-1, -3, -4, MAGEC1, BAGE, XAGE3, NY-ESO1, SSX2, SCP1, and PRAME1. Two genetic loci GAPDH and GUSB were used as reference ones. Specific oligonucleotide primers (Table 1) developed by Vodolazhskiy et al. were used [4, 7].
The obtained cDNA was amplified in 20 µl of the solution that contained 12 ng of cDNA matrix, 0.2 mM dNTPs, 2.5 mM MgCl2, 1× PCR buffer and 2 units of active SynTaq DNA-polymerase, EVA-Green stain, and 612 nM of direct and reverse primers for the reference genes or the target gene. RT-PCR-amplification was performed using thermocycler “Bio-Rad CFX96” (Bio-Rad, USA) by the following protocol: primary denaturation: t=95 °C for 240 s; 40 cycles: t=95 °C for 10 s, t=58 °C for 30 s, t=72 °C for 30 s.
The relative expression (RE) was calculated by the formula RE=2-ΔΔCt [4, 11, 12]. The normalization of the results was performed by two reference genes (GAPDH and GUSB) and the level of expression of the respective target genes in the samples of normal tissues by the following protocol:
1. Normalization by the mean of the reference genes: ΔC(t) = C(t)target – C(t)reference.
2. Calculation of the median ΔC(t) for each gene for the control (relatively healthy) and test (tumor) groups.
3. Normalization by the control group: ΔΔC(t) = ΔC(t) Median of the test group – ΔC(t) Median of the control group.
4. Final result (fold difference)): 2-ΔΔC(t).[4].
The statistical analysis was performed using the applied statistical software package Microsoft Excel 2013 (Microsoft Corporation, USA) and STATISTICA 8.0 (StatSoft Inc., USA). The patients were divided into two independent groups: Group A – with regional metastases (10 patients) and Group B – without regional metastases (14 patients). The statistical significance was defined by the Mann-Whitney non-parametric test. The null hypothesis on the absence of differences was rejected at p<0.05.
Table 1
A panel of primers for the evaluation of the relative expression of the genes [4, 7]
|
No. |
Genetic locus |
Primer sequence |
|
|
Direct |
Reverse |
||
|
1 |
GAPDH |
GTCAAGGCTGAGAACGGGAA |
TCGCCCCACTTGATTTTGGA |
|
2 |
GUSB |
CAGGACCTGCGCACAAGAC |
CTAGCGTGTCGACCCCATTG |
|
3 |
MAGEA1 |
GAAGGAACCTGATCCAGGC |
AGGGAATTCTGTCCTCTGGG |
|
4 |
MAGEA2 |
CGAAGGCTCCGTGAGGA |
CTGTATTGACCTGAGTCACC |
|
5 |
MAGEA3 |
TGAGCAAAGAGCGACGG |
TCAGACTGTCCCCTCAGAA |
|
6 |
MAGEB1 |
TTCAGTGTGGTGTCCAGCAA |
CGAGTTGTACTCCTGGATGATCA |
|
7 |
MAGEB2 |
AGCCAGGGGTGAATTCTCTG |
GGCACGGAGCTTACTCTCCT |
|
8 |
GAGE1 |
CTGATGGGCACGAGATGGAC |
CCAGTCTCGGCAACATAGTGA |
|
9 |
GAGE3 |
TCACACAGCTGAGTTGGCGA |
CTGTGTGAAATATGAGTTGGCGC |
|
10 |
GAGE4 |
GAGGAGGTGAAAACGCCTGG |
GCATCATTTCAACGTGCCTTCG |
|
11 |
MAGEC1 |
ACGAGGATCGTCTCAGGTCC |
CCAGGTCTTCAACTCCTGCC |
|
12 |
MAGEA4 |
CTGACCAGCAGCTTGGGATC |
TCCAGGGAATCCTGTCCTCCT |
|
13 |
BAGE |
GCCGGCTCCTTTCAGGATTT |
ACATCTTTCAGGAGCTTGGTCAC |
|
14 |
XAGE3 |
ACTTGCCCTGAGACTTAGTT |
ACTTGCCCTGAGACTTAGTTT |
|
15 |
NY-ESO1 |
GAGTTCACTGTGTCCGGCAC |
TGGAGACAGGAGCTGATGGA |
|
16 |
SSX2 |
TACGGTTGGTGCTCAAATACC |
CCGAGGCTTTCATCTTTTCCT |
|
17 |
SCP1 |
AGGTGAAACCTCAGACCCT |
AGTCTTTGCAAATGGAAACTCAAT |
|
18 |
PRAME1 |
GCTGAGCCATTGTCTCGTTACT |
AGGTCTCAGTCACTTGTTGCC |
RESULTS
The study of the combined sampling consisting of 24 patients with metastatic (T1-3N1-2M0) and non-metastatic (T1-3N0M0) cancer revealed a statistically significant (p<0.05) increase in the expression of two C/T genes SSX2 and PRAME1 by 1.8 (in 25% of patients) and 2.9 (in 50% of patients) times, respectively, in the tumor tissue in comparison with normal tissue and a decrease in the expression of one C/T gene BAGE by 2.4 times (in 20% of patients) in the tumor tissue in comparison with normal colon tissue (Figure 1). The expression of the other 13 genetic loci MAGEA1, MAGEA2, MAGEA3, MAGEB1, MAGEB2, GAGE1, GAGE3, GAGE4, MAGEC1, MAGEA4, XAGE3, NY-ESO1 and SCP1 in the tumor tissue did not differ significantly from the level of expression in the normal colon tissue (p>0.05).
Patients with non-metastatic colorectal cancer (T1-3N0M0) had a statistically significant increase (p<0.05) in the expression of C/T genes MAGEB1 (in 40% of patients), SSX2 (in 60% of patients) and SCP1 (in 50% of patients) by 2.2 times, 2.2 times, and 2.7 times, respectively, in the tumor colon tissue in comparison with normal colon tissue and a decrease in the expression of the gene BAGE by 1.9 times (in 40% of patients) in the tumor tissue in comparison with normal colon tissue (Figure 2).
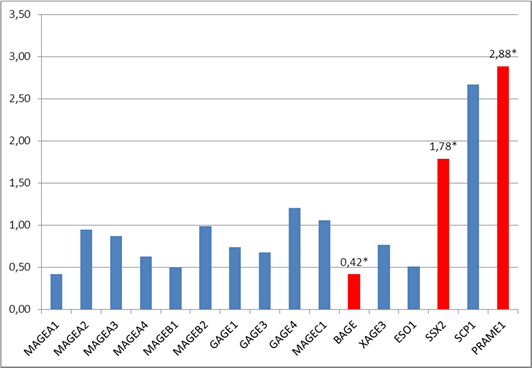
Fig 1. The ratio of the expression of C/T genes in the tumor tissue of the rectum in comparison with normal tissue (combined sampling, n=24), * – statistically significant difference (p<0.05) between tumor and normal tissues
Patients with regional metastases (T1-3N1-2M0) had a statistically significant (p<0.05) increase in the expression of the genes GAGE1 (in 40% of patients), SCP1 (in 70% of patients) and PRAME1 (in 40% of patients) by 2.5 times, 9.1 times, and 1.9 times, respectively, in the tumor colon tissue in comparison with normal colon tissue and a decrease in the expression of the genes MAGEA2 (in 40% of patients), MAGEB1 (in 50% of patients), MAGEB2 (in 70% of patients), GAGE4 (in 40% of patients) and NY-ESO1 (in 40% of patients) by 2.7, 3.0, 5.9, 3.1 and 3.3 times, respectively, in the tumor colon tissue in comparison with normal colon tissue (Figure 2).
It should be mentioned that in the group of patients with metastatic cancer (T1-3N1-2M0), the expression of the genes GAGE1 and SCP1 in the tumor tissue was 3.4 times higher (p<0.05) than the level of expression of these genes in the group of patients with non-metastatic cancer (T1-3N0M0). At the same time, in the group of patients with metastatic cancer, the expression of the genes MAGEA2, MAGEB1, MAGEB2, GAGE3, GAGE4, NY- ESO1 and SSX2 in the tumor tissue was significantly lower (p<0.05) by 2.6; 6.8; 5.9; 2.2; 3.1; 4.5 and 3.2 times, respectively, than in the group of patients with non-metastatic cancer (Figure 2).
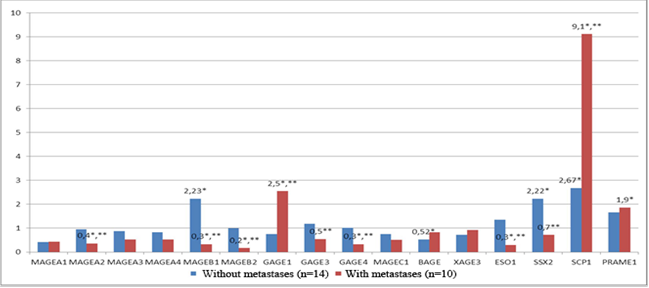
Fig 2. The level of transcriptional activity of C/T genes in the tumor tissue of the rectum in comparison with the normal tissue in patients with metastatic (n=10) and non-metastatic cancer (n=14). * – statistically significant differences (p<0.05) between tumor and normal tissues, ** – statistically significant differences (p<0.05) between the patients with metastatic and non-metastatic cancer.
The obtained data showed that patients with metastatic and non-metastatic colorectal cancer had significantly different transcriptional pr of CTA. In both groups, the authors observed an increased expression of CTA of different classes: CT-X (MAGEB1, SSX2, GAGE1) and non-X(SCP1, PRAME1), testicular selective (SSX2, GAGE1, SCP1) and testicular limited (MAGEB1, PRAME1). However, only the tumor tissue obtained from patients with metastatic cancer showed a significant decrease in the expression of some CTA genes in comparison with patients with non-metastatic cancer and normal tissue.
An increased expression of the genes SSX2 and GAGE1 and a decreased expression of the genes MAGEB1, MAGEB2, GAGE3, GAGE4, and NY-ESO1 agree with the results of other studies [1, 9]. Unlike the data presented in [13, 14] for CTA genes from MAGEA family, the present study did not reveal a statistically significant increase in the transcriptional activity.
CONCLUSIONS
The results of the present study revealed a differentiated transcriptional activity of C/T genes in patients with metastatic and non-metastatic colorectal cancer that was observed in the change in the level of expression of genetic loci MAGEB1, SSX2, BAGE and SCP1 (for non-metastatic colorectal cancer) and GAGE1, SCP1, MAGEA2, MAGEB1, MAGEB2, GAGE4 and NY-ESO1 (for metastatic colorectal cancer). The obtained data provided grounds for further studies on the expression of C/T genes on larger samplings for the formation of a panel of effective immune therapeutic targets for two groups of patients: with regional metastatic (T1-3N1-2M0) and non-metastatic (T1-3N0M0) colorectal cancer. Genetic loci MAGEB1, MAGEB2, SSX2, BAGE, SCP1, GAGE1, 3, 4, MAGEA2 and NY-ESO proved to have high potential in the prognosis of the development of regional metastases in patients with colorectal cancer.
FINANCIAL SUPPORT AND SPONSORSHIP
Nil.
CONFLICTS OF INTEREST
The authors declare no conflict of interest



